20.12.2023
4 minutes of reading
Researchers are using genetic engineering to improve their understanding of the living world and inject this newly acquired knowledge into industrial biotechnologies in order to decarbonize the energy and chemicals sectors. It is within this context that IFPEN’s biotech teams are conducting research aimed at improving the efficiency of enzymes in the conversion of plant cellulose and other polysaccharides into fermentable sugars. It was research of this type, for example, that helped establish the FuturolTM process as a cutting-edge technology with a bright future. To further advance this global understanding of the living world, teams are also now using complex algorithmic processing to interpret scattered and incomplete experimental data. Below we present an analogy to set the scene and explore this new world.

The living world: let the inquest begin
In a court room, a trial is being held behind closed doors. Is it possible, without actually being there, to understand the evidence that led to the verdict pronounced? We only have partial, retrospective information on what happened during the hearing, such as the identity and function of some of the players involved (plaintiff, defendant, lawyer, prosecutor, judges, clerks), but not all (jury, witnesses). In addition, we may have access to charges, courtroom sketches, excerpts from court minutes, radio or TV reports, lawyers' statements, etc. How, then, can we reconcile all these different types of data (written documents, drawings, voices, photos, videos) to obtain a coherent vision of the proceedings? To do so would also require “smart” tools, capable of analyzing these fragmented and complementary pieces of evidence.
AI tools that are powerful yet often restricted to a single modality: sound, image, etc.
Artificial intelligence has been making the headlines recently and is now familiar to the public at large: ChatGPT, Siri, MidJourney, Dall-E, Google Bard, Mubert, Bing AI,… These tools exploit a particular modality of data available in very large quantities on the Internet, be it textual documents, recorded voices, sets of images or video extracts, etc. So-called “artificial intelligence” algorithms are used to design learning models that meet a very specific objective: generating text or images, classifying, and so on. To achieve this, these algorithms strive to capture the basic building blocks of data. These building blocks are abstract and can often not be interpreted by human beings. However, their subsequent reorganization makes it possible to imitate certain tasks, known as intelligent tasks, which conscious species are able to produce within the confines of their nervous systems: for example, answering written questions, giving the illusion of a conversation, creating realistic images from a simple description, faking a video.
Ideal intelligence needs to exploit several modalities
To explore this further, it seems logical to seek to use the different modalities available for learning data in conjunction with each other. The challenges involved in such an approach are obviously considerable, ranging from the ever-increasing volume of data through to the combination of relevant associations. Research is emerging on this topic, such as SeamlessM4T (Massively Multilingual & Multimodal Machine Translation) developed by Meta (formerly Facebook) to integrate vision and language. Returning to our courtroom analogy, such tools could assemble a multimodal puzzle of the trial, from all the partial data available, from the textual announcement of the verdict by the media to the non-verbal language of the players involved in front of the cameras, to the pronouncement of the verdict.
An example of multimodality: omics data
Away from the courtroom, IFPEN faces similar questions. However, they concern fragmented and complementary scientific data, which need to be processed by intelligent tools. But for what purpose, and to understand what verdict?
In a particular biological context related to the energy transition, IFPEN is conducting research in the field of bio-based chemistry and biofuels with a view to optimizing specific biotech processes. These processes depend on the use of microorganisms, whose behavior needs to be fully understood in order to make them as efficient as possible. This new knowledge would make it possible to improve the performance of these microorganisms, increase the yields of these biotech processes, reduce their cost and promote their deployment.
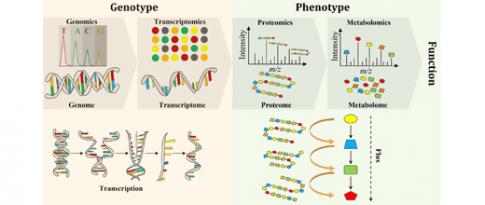
For a particular cell, we can understand some of the genes and their functions, as well as some of their actions and interactions. And we can also access other important pieces of information by collecting so-called "omics" data: genomics to study genes, transcriptomics to measure gene expression, epigenetics to understand how environment may influence gene expression without alteration of the genome, etc. Like in the courtroom scenario, these scattered omics data each provide a partial view of the molecular phenomena at play within a “cell” that result in protein production.
IFPEN and CentraleSupélec/INRIA investigate through PhD research
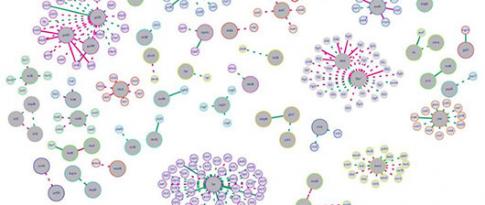
Experts from IFPEN and the OPIS project team at CentraleSupélec/INRIA joined forces for a PhD thesis aimed at developing new methods for analyzing the behavior of microorganisms [1]. A systemic approach (figure 1) was adopted to take into account different omics data modalities : genomics, transcriptomics, metabolomics, epigenomics, etc. Although heterogeneous in nature, these data can be represented in graph form. The resulting interaction networks, themselves interacting with each other, carry complementary information useful for the understanding of the underlying biological mechanisms. Two principal machine learning techniques on large-dimension graphs were developed, retrospectively called BRANEnet [2] and BRANEmf [3]. These techniques adapted to the analysis of multi-omics data draw on recently developed numerical methods for analyzing text and social networks. They make use of statistical and algebraic methods, as well as machine learning. Also known as embedding methods, they convert graphs into new numerical representations of reduced dimensions, which are easier to manipulate by an algorithm. Their application to graphs made up of different omics modalities makes it possible to grasp the main relationships between genes. This simplified description of the way a cell works is then used to carry out various tasks of interest for the understanding of biological phenomena: grouping of genes associated with the same molecular function, prediction of molecular interaction, elucidation of inter-gene dependency, etc.

A case study: brewer’s yeast
Embedding methods have been validated for well-known model biological organisms such as Saccharomyces cerevisiae, the famous brewer’s yeast, by comparison with the state of the art. They thus offer new avenues for understanding molecular mechanisms, in order to improve the production of molecules and bio-based products in the future.
Open-source algorithms and data
In a spirit of sharing and open science, all the data, algorithms and computer codes associated with this research are made freely available to the public and the scientific community1.
1 Associated codes:
- BraneNET
- BraneMF
References:
[1] Multilayer Graph Embeddings for Omics Data Integration in Bioinformatics, Surabhi Jagtap, Thèse de doctorat en Informatique mathématique, soutenue le 02 février 2023, Université Paris-Saclay.
[2] BRANEnet: Embedding Multilayer Networks for Omics Data Integration. Surabhi Jagtap, Aurélie Pirayre, Frédérique Bidard, Laurent Duval, Fragkiskos D. Malliaros, BMC Bioinformatics, 2022
[3] BraneMF: Integration of Biological Networks for Functional Analysis of Proteins. Surabhi Jagtap, Abdulkadir Çelikkanat, Aurélie Pirayre, Frédérique Bidard, Laurent Duval, Fragkiskos D. Malliaros, Bioinformatics, 2022
Scientific contacts : Aurélie Chataignon Pirayre, Laurent Duval, Frédérique Bidard (IFPEN), Fragkiskos Malliaros (OPIS)
You may also be interested in
‘‘BRANE Power’’: of genes and algorithms, an alliance for green chemistry
THESIS BY AURÉLIE PIRAYRE, 2018 YVES CHAUVIN PRIZE
The “Omics”, seven hired hands working for biotechnology
Most frequently, the use of a microorganism in the production of biofuels or biosourced chemicals requires its optimisation. This is achieved through genetic engineering, which involves inactivation and/or addition of one or more genes, to improve the ability of this microorganism to produce a target molecule. (...) At IFPEN, access to genomics (DNA) has helped in gathering data on the composition and structure of the genomes of the microorganisms...